CSC News
Exploring Deep Learning AI for Cancer Treatment
Researchers at NC State and UNC-Chapel Hill are exploring how to potentially use AI to match anti-cancer drugs to a patient’s specific DNA.
Their research takes a precision medicine approach by using data-centered tools and methods to optimize treatment by tailoring it to a patient’s specific health conditions and needs.
The proposed AI model demonstrates the potential to predict how different types of cancer would respond to experimental drug compounds. This way, the most promising anti-cancer drugs would be identified early in the development process, reducing unnecessary experimental costs and time and accelerating the development of new treatments.
Leading the preliminary research is Zhishan Guo, an associate professor in the Department of Computer Science, and Ning Sui, a molecular biochemist and assistant professor in the Department of Molecular and Structural Biochemistry. The team is also collaborating with Dr. Young E. Whang, an associate professor of medicine and pathology and urology at the University of North Carolina at Chapel Hill.
The research project was supported by a pilot grant award from the North Carolina Translational and Clinical Sciences (TraCS) Institute. The NC TraCS Institute combines the research strengths, resources and opportunities of UNC-Chapel Hill with those of its partner universities, NC State and North Carolina A&T. The TraCS’ pilot grant program aims to advance research that improves how medical practices and principles translate into more effective care.
Beyond a One-Size-Fits-All Approach to Treating Cancer
Cancer is the second-leading cause of death in the United States, with mortality rates approaching the high rates of heart disease. Traditional anti-cancer drug treatment often adopts a “one-size-fits-all” approach.
For instance, two patients can have the same type of cancer, but the tumors differ at the genetic level. “There are tiny details in genomes that can actually make a big difference in a patient’s drug response to a particular medication,” said Sui, the molecular biochemist of the interdisciplinary research team.
With a heterogeneous disease like cancer, reactions to medication can be highly variable and unpredictable. “So that justifies individualized, personalized medicine — because humans are so different,” Sui said.
Guo’s and Sui’s research pilot explored individual genetic differences in cancer cells grown in labs at the molecular level. Then, they combined this information with the molecular structure of existing anti-cancer drugs to predict treatment responses in the lab-grown cells.
“Instead of guessing which drug might work, doctors can choose medications specifically tailored to each patient’s genetic profile, giving them the best chance of successful treatment,” said Yuhan Zhao, a Ph.D. student in computer science participating in this interdisciplinary pilot project.
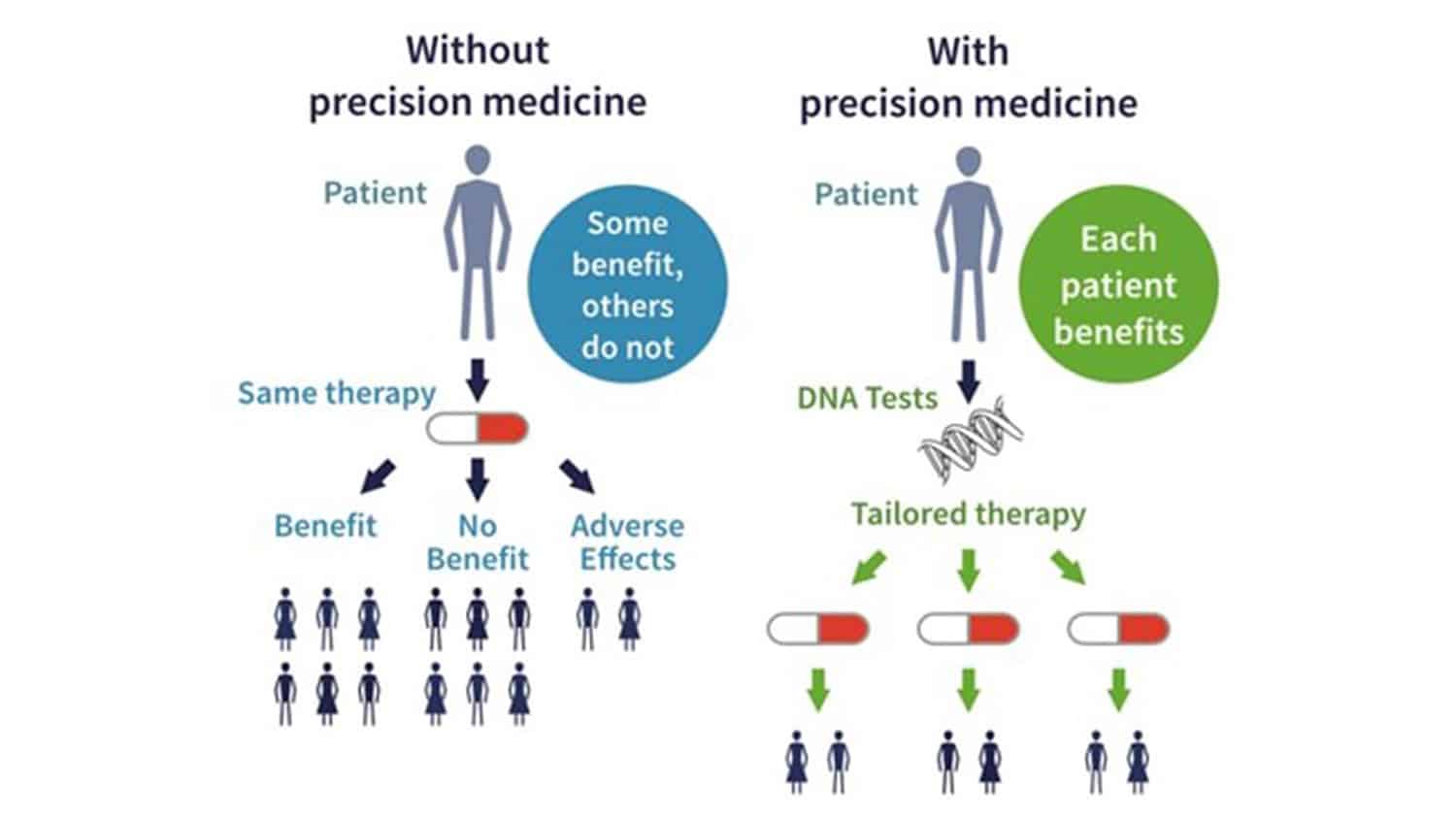 The left side of the graphic illustrates traditional medicine’s “one-size-fits-all” approach to cancer therapy, resulting in varying patient responses. The treatment benefits some, while others may see no effects, and a third group may experience adverse effects. To the right, a precision medicine approach provides more effective and tailored treatment, using DNA testing to match anti-cancer drugs to a patient’s genetic profile.
The left side of the graphic illustrates traditional medicine’s “one-size-fits-all” approach to cancer therapy, resulting in varying patient responses. The treatment benefits some, while others may see no effects, and a third group may experience adverse effects. To the right, a precision medicine approach provides more effective and tailored treatment, using DNA testing to match anti-cancer drugs to a patient’s genetic profile.
Currently, doctors use DNA testing for cancer treatment only in specific cases. With well-studied cancers like lung cancer, doctors use DNA sequencing for some specific mutations to discern which medication would be more effective according to existing cancer research. For cancers with fewer known characteristics or patterns, comprehensive DNA sequencing is less common.
The team’s research explores the potential benefits of using DNA testing to improve the matching of anti-cancer medications to patients — a goal that is becoming increasingly realistic as sequencing becomes more affordable. Their current AI model is based on data from lab-grown cancer cell lines, serving as a proof of concept for future patient-specific applications.
“DNA sequencing is so inexpensive nowadays, as long as we have a way to process the sequencing data efficiently, it’s really a very low-cost thing compared with the benefit it can bring,” Sui said.
Making Different Data Speak the ‘Same Language’
One challenge has been making two differing types of data “talk to each other.” Genetic information and the chemical data of medications are structured differently, and one does not easily translate to the other.
“We needed to find a way for the computer to ‘understand’ both languages and integrate them effectively,” said Zhao.
The team developed an advanced, deep learning AI model to process these different kinds of data — or, in other words, “speak the same language.” For 782 different cancer cells, the team processed nearly 30,000 genes of molecular data for each one. Interweaving that data with approximately 250 different drugs, they created a training set of over 160,000 cell-drug pairs that can carry out experimental research. This preliminary deep learning model demonstrates the potential to predict how a patient’s specific genetic profile would respond to 256 existing medications.
What’s the difference between machine learning and deep learning?
In summary, machine learning and deep learning, both subsets of artificial intelligence (AI), differ in how they are trained and the level of complexity of their problem-solving capabilities.
But, before explaining further, it helps to review a few general concepts about computing. Classical computing, or traditional programming, involves explicitly coding a computer with a fixed set of sequential instructions (algorithms) to perform specific tasks, much like following a step-by-step recipe in a cookbook. Its process is predetermined, and it yields a predefined output.
Instead of being programmed for specific scenarios, a computer using AI algorithms can identify patterns. A common type of AI, machine learning enables a computer to learn from these patterns, allowing it to solve problems and make informed decisions or predictions. Machine learning typically requires human expertise to identify and extract features — the useful parts of raw data —which are then used to train the algorithm. Generally simpler, these algorithms have fewer layers of computation.
Deep learning is a subfield of machine learning that uses a multi-layered model called a neural network — a reference to the way it mimics how the human brain’s biological neurons work together to identify phenomena, weigh options and arrive at conclusions. What makes deep learning powerful is that it can directly process raw, unstructured data, such as images, text and audio, without requiring human feature engineering. The data is processed through multiple layers — often referred to as architectures — allowing the neural network to learn increasingly abstract features. This complexity requires very large datasets and strong computing power for effective training.
So, to use an analogy: If classical computing is like using a cookbook — you follow prewritten instructions precisely and the outcome is a specific dish — AI is like having a knowledgeable chef’s assistant at your disposal. This assistant isn’t just programmed for one recipe; it can access broad culinary knowledge and apply reasoning to answer your questions about any dish or cooking in general.
Machine learning enhances this chef’s assistant by making it adaptive. If you express a dislike for an ingredient or praise a dish, it learns from your feedback and adjusts future recommendations, personalizing suggestions over time.
Deep learning takes it a step further: the adaptive assistant becomes an intuitive master chef. This culinary expert has absorbed vast amounts of experience, understanding subtle nuances of abstract features like taste and texture. Beyond learning your preferences, it would anticipate your needs by detecting less obvious patterns — such as your mood or the time of year — to create innovative, highly attuned menus from scratch. This is possible because deep learning builds a rich, multi-layered internal model of highly specific and diverse data, mimicking human-like intelligence in its ability to synthesize complex information.
Using a deep learning AI model also helped the research team overcome another challenge: the lack of existing data on molecular information for both human DNA and anti-cancer drugs. Generally, deep learning models are especially effective at processing long data sequences such as DNA strands or the molecular structures of drugs.
Guo said that the team experimented with several different machine learning models and techniques until they discovered a special component of deep learning that can capture long-range dependencies, providing guidance on how certain genes and mutations will respond to a drug over time.
“It’s quite a new technique,” said Guo, who is also a co-founder of NC State’s Cyber Physical Systems group. “I believe that’s the first time it’s being used for this drug response prediction.”
Potential Advances in Anti-Cancer Treatment
Compared to current methods, Guo’s and Sui’s AI model outperforms existing approaches. The research team’s methods can more accurately predict drug efficacy, making it promising for applications in anti-cancer drug development and personalized therapy.
“The key outcome for our one-year project was that we were able to design and verify a machine learning-based system that can take gene expression and mutation information, as well as cancer drug structure information — and with 98% precision — predict whether a drug is effective for certain types of gene expression and mutations,” said Guo. “This is really something that’s novel and exciting.”
Future iterations of the research team’s deep learning model could incorporate electronic health records and hospital-based genetic testing results, providing doctors with a powerful, cost-effective and collaborative AI tool to guide anti-cancer treatment decisions.
Adapted for the NC State Department of Computer Science, original article by H'Rina DeTroy, NC State News.
Return To News Homepage